Integrated Genomics Laboratory Services Overview
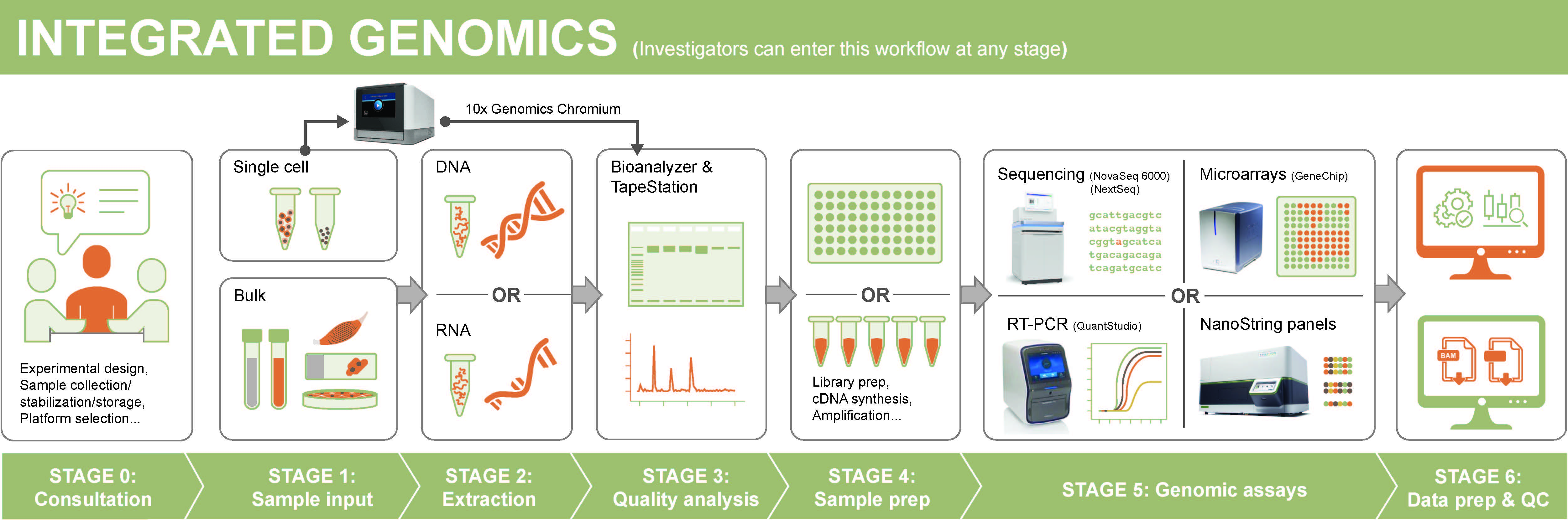
We provide a coordinated pipeline of services which can be accessed at whatever point the researcher requires. Our services include in depth consultation, nucleic acid extraction, bulk and single cell sequencing, targeted gene analyses, and data processing and QC.
Services








