CEDAR Jobs

We are building a team of the best and brightest minds to help us end cancer as we know it.
We are looking for outstanding researchers to help us end cancer as we know it. We encourage high-risk, high-reward research projects because defeating cancer requires unconventional thinking. Our research is rooted in team science and collaboration; we encourage ideas from everyone.
Why work at CEDAR?
As a CEDAR researcher, you are part of a team of scientists at the OHSU Knight Cancer Institute, one of the nation’s foremost centers for cancer research. We are pioneers in targeted therapy and early detection. We offer unmatched opportunities to accelerate your career.
At CEDAR, you can:
- Grow as a scientist or researcher
- Find great mentors
- Develop your skills
Grow as an independent scientist
- Perform cutting-edge science. Our projects range from understanding basic cancer biology to developing new technologies for detection, computational analysis or therapeutic intervention. Learn about some of our research programs.
- Choose your own path by finding your own collaborators, joining other projects, submitting your own proposals and overseeing your own research projects. CEDAR supports the startup approach of patent development and fail-fast projects alongside the traditional approach of manuscript writing and publishing.
- Work alongside the team from SMMART, the Knight Cancer Institute's unique clinical trials platform for precision medicine. Make a new molecule or repurpose an existing one, and see it tested in the clinic by a multidisciplinary team you helped build.
- Speed your path to independence. CEDAR offers generous internal funding to researchers, including scientists and postdoctoral researchers.
Find great mentors
Our scientists and faculty include some of the world's leading experts in early detection. At CEDAR, every researcher, scientist, and postdoc is matched with an experienced supervisor to help develop a research portfolio, consider career plans, and tailor professional development.
Get professional development
Professional development is essential the growth and success of our researchers. All CEDAR staff receive:
- Annual funding to support travel, conference activities, and continuing education
- Professional development funding to present research and network, and to explore techniques and ideas to advance CEDAR's mission and guiding principles.
For postdoctoral researchers, we offer exceptional opportunities for leadership, development, and funding:
- The OHSU Office of Postdoctoral Affairs offers a wealth of resources to postdocs. Its mission is to support your professional development for all stages of your career.
- The OHSU Training Future Faculty Program supports teaching excellence by training postdocs through workshops, peer coaching, and professional development.
Earn a competitive salary with annual increases
All full-time CEDAR employees receive annual increases. First-year postdocs earn a salary that exceeds the minimums set by the National Institutes of Health.
Plug into our network
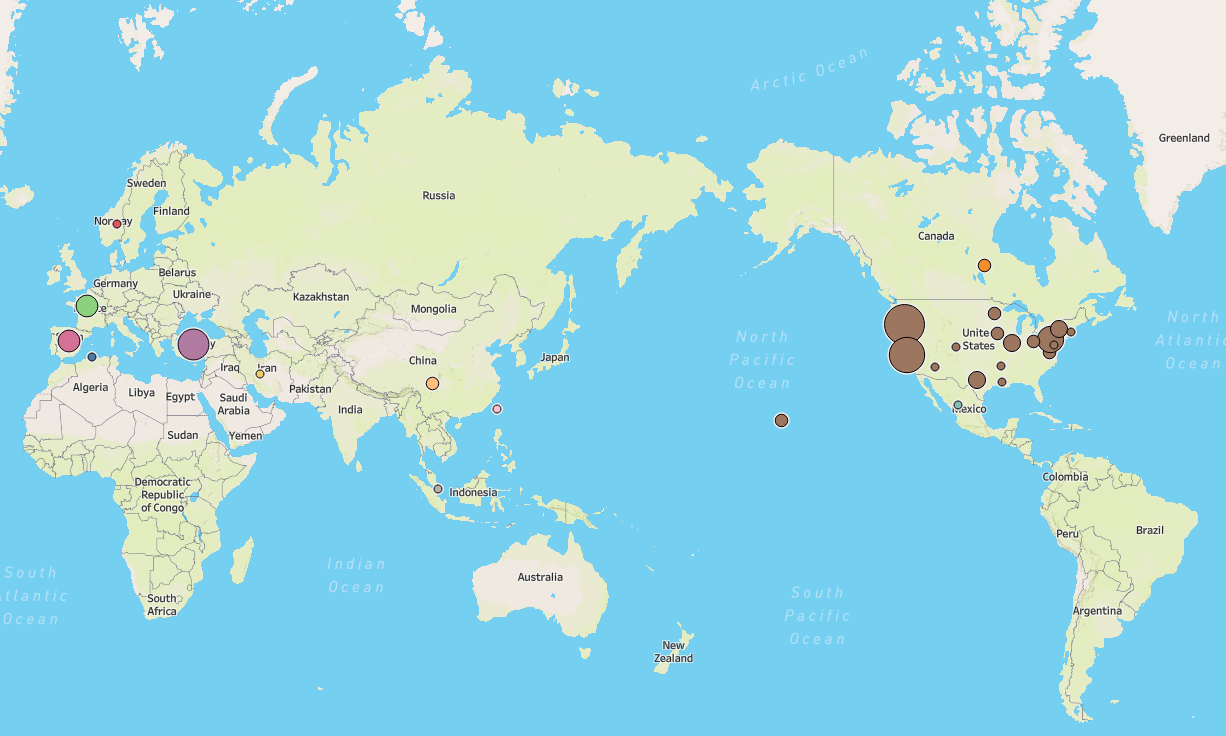
CEDAR draws top researchers from all over the world, with more than 20 countries represented amongst our staff. Click the map below to learn more. We also have a far-flung alumni network of researchers.
Become a graduate researcher
We recruit graduate researchers from two OHSU graduate programs:

