Intracellular Paired Agent Imaging Enables Assessment of Drug Biodistribution & Therapeutic Efficacy
How is drug efficacy currently measured and why isn't it predictive of lasting response?
For a therapeutic to be effective, it must engage with its drug target in the complex setting of the diseased tissue, where successful therapy is dictated by the duration, completeness and cellular heterogeneity of the drug engagement with its target. A major contributing factor to therapeutic failure is insufficient drug target engagement and off-target activity, which are typically assessed by bulk sampling of plasma or tissues yielding a heterogeneous average not representative of cell-to-cell drug distribution, target binding and off-target effects. A variety of methods to measure available drug distribution and drug target engagement have been developed, however, they have not yet been able to reliably predict therapeutic efficacy largely because they cannot measure on a cell-by-cell basis to understand efficacy in heterogeneous diseases such as cancer. A variety of small molecule drugs with labeling modifications have been generated including fluorescent, biotin and photoclickable labels, to facilitate direct visualization of tissue distribution and target engagement, pull-down assays and imaging. Conversely, drug target proteins have been genetically modified enabling visualization. While useful to provide initial spatial assessment of drug and target distribution, modifications can vastly alter drug distribution or target engagement. To overcome this difficulty, a number of label-free methods have been developed including mass spectroscopy imaging, assessment of drug targets in tissue lysates, cellular thermal shift assay and positron emission tomography imaging using radiolabeled drugs as a read out following administration of the parent drug. While the difficulties of labeled parent drug or target are avoided, these approaches do not enable assessment of untargeted drug accumulation and lack single cell resolution.
What is intracellular paired agent imaging (iPAI) and will it accurately predict therapeutic efficacy?
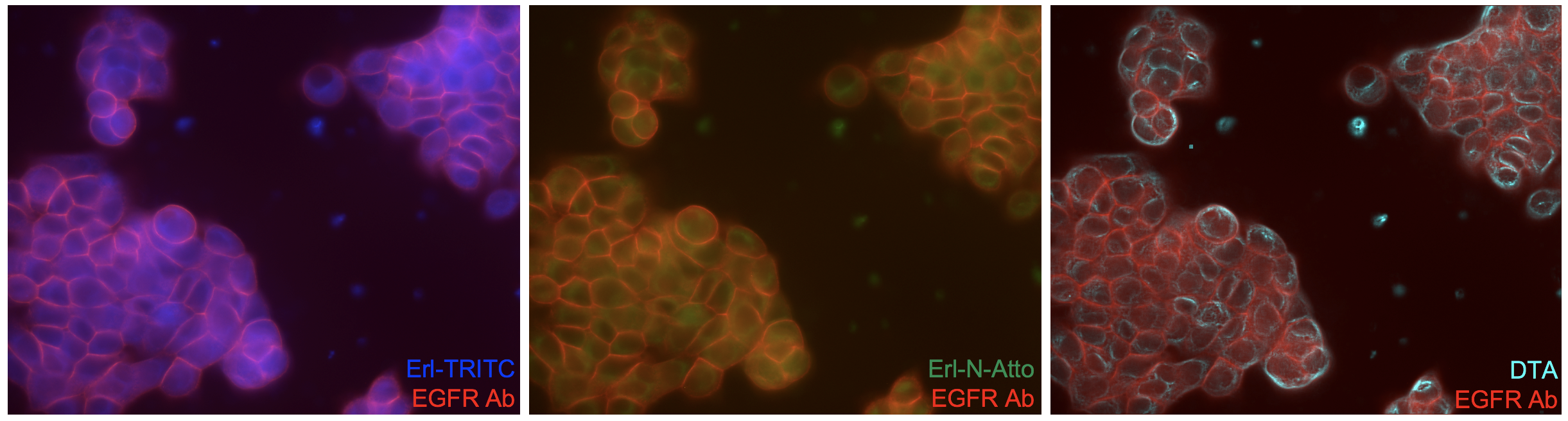
Quantification of available drug targets necessitates accounting for both the drug that binds to its target as well as the drug that accumulates in the cells and tissues in a non-specific or untargeted manner. While targeted uptake is the goal of drug development, all drugs have some degree of untargeted accumulation due to drug affinity, biodistribution, pharmacokinetics and metabolism, which can vary on a cell-by-cell basis in heterogeneous diseases such as cancer. To facilitate quantification of cell-to-cell variation in tissues, we have adapted a technique from nuclear medicine termed intracellular Paired Agent Imaging (iPAI), in which receptor mediated imaging was made quantitative by co-administering a radiolabeled receptor targeted antibody and an untargeted antibody with a different radiolabel. The uptake of the untargeted antibody was then used to estimate the extent of non-specific uptake yielding a quantitative assessment of receptor density. This technique has been reinvigorated by the optics community and used extensively by our group and others, where spectrally distinct targeted and untargeted imaging agents are used to correct for untargeted uptake, thus providing quantitative in vivo or ex vivo assessment of receptor density for molecular imaging. We have extended the iPAI technique to small molecule therapeutics using spectrally distinct fluorescently labeled targeted and untargeted drugs to map drug target availability (DTA), which accurately predict therapeutic efficacy.
iPAI Figure
Acknowledgements
This work was funded by the V Foundation for Cancer Research and the Collins Medical Trust.