Our mission: enabling epigenetics research
Our mission is to promote and facilitate epigenetics research and education across students, faculty, and physicians. The KCVI Epigenetics Consortium is directed by Dr. Lucia Carbone and includes several investigators whose research involves various aspects of epigenetics (e.g., environmental epigenetics, cancer, maternal reprogramming, neuroscience, reproductive biology).
In order to facilitate epigenetics research, the Epigenetics Consortium includes a service core. This service core functions on a fee-for-service basis, with a sample-to-data model in mind.
Investigators inside and outside of OHSU can choose our wet-lab services, dry-lab services, or both. The service core provides end-to-end support for a number of DNA methylation and chromatin analyses. These services include, but are not limited to, Bisulfite DNA sequencing and HiC.
Life cycle of a project
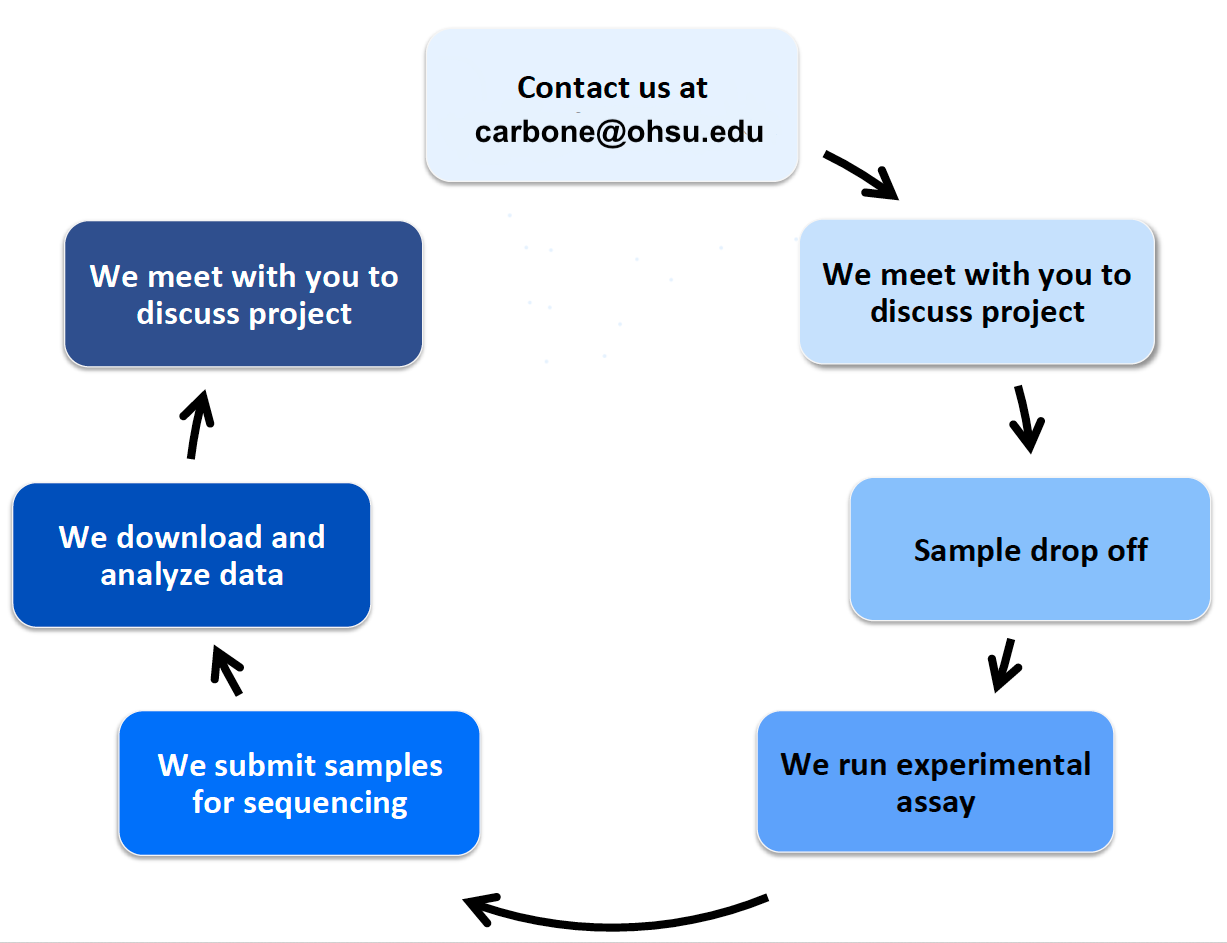
Contact us
If you are interested in any of our services, please fill out the project inquiry form and send it to us here
We look forward to talking with you.
Locations
