Skalsky Laboratory
Viruses utilize various strategies to manipulate gene expression throughout infection, thereby combating host anti-viral defense mechanisms and enabling pathogen success. Research in the Skalsky lab examines the interface between oncogenic herpesviruses and the host RNA interference machinery in order to understand how post-transcriptional interactions such as these contribute to the viral life cycle, virus-associated autoimmune disease, and virus-driven cancers.
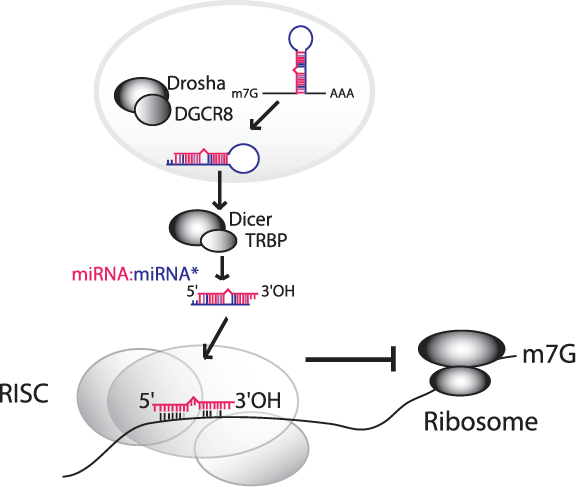
An estimated 15% of human cancers are associated with viral infection. Members of the gamma-herpesvirus family, such as EBV and KSHV, are linked to AIDS-associated non-Hodgkin's lymphomas and other B cell malignancies. The molecular mechanisms driving lymphomagenesis are not well defined, presenting a major challenge in treating these diseases.
A major focus of the lab is to explore the roles of microRNAs in viral pathogenesis. miRNAs are a class of small, non-coding RNAs that post-transcriptionally regulate gene expression. Due to the nature of their regulatory influence, miRNAs represent attractive therapeutic targets. Encoded by all metazoans, miRNAs have also been identified in many herpesviruses. One goal of the lab is to identify miRNA targets and/or targeted pathways that are critically involved gamma-herpesvirus pathogenesis. We use combined high-throughput biochemical and bioinformatics-based approaches to interrogate miRNA targets and miRNA regulated pathways during viral infection.
Postdoctoral Fellows: Postdoctoral candidates with expertise in virology, RNA biology, and/or bioinformatics are encouraged to apply and should submit a short cover letter, three references, and copy of their CV to Dr. Skalsky.
Undergraduates: Motivated, bright young individuals with interest in virology and interested in pursuing a career in biomedical research are encouraged to contact Dr. Skalsky about student positions in the lab.
Job opportunities: Please check the OHSU Human Resources website for current openings.
-
Chen Y, Fachko DN, Ivanov NS, Skalsky RL. B Cell Receptor-Responsive miR-141 Enhances Epstein-Barr Virus Lytic Cycle via FOXO3 Inhibition. (2021) mSphere 6(2):e00093-21
-
Bouvet M, Voigt S, Tagawa T, Albanese M, Chen YA, Chen Y, Fachko DN, Pich D, Göbel C, Skalsky RL, Hammerschmidt W. Multiple Viral microRNAs Regulate Interferon Release and Signaling Early during Infection with Epstein-Barr Virus. (2021) mBio 12(2):e03440-20
-
Diggins NL, Crawford LB, Struthers HM, Hook LM, Landais I, Skalsky RL, Hancock MH.
Techniques for Characterizing Cytomegalovirus-Encoded miRNAs. (2021) Methods Mol Biol. 2244:301-342 -
Okoye AA, DeGottardi MQ, Fukazawa Y, Vaidya M, Abana CO, Konfe AL, Fachko DN, Duell DM, Li H, Lum R, Gao L, Park BS, Skalsky RL, Lewis AD, Axthelm MK, Lifson JD, Wong SW, Picker LJ. Role of IL-15 Signaling in the Pathogenesis of Simian Immunodeficiency Virus Infection in Rhesus Macaques. (2019) J Immunol. 203(11):2928-2943
-
Chen Y, Fachko D, Ivanov NS, Skinner CM, Skalsky RL. Epstein-Barr virus microRNAs regulate B cell receptor signal transduction and lytic reactivation. (2019) PLoS Pathogens. 15(1):e1007535
-
Skinner C.M., Ivanov N., Barr S.A.,Chen Y., and R.L. Skalsky. An Epstein-Barr Virus microRNA Blocks Interleukin-1 (IL-1) Signaling By Targeting the Interleukin-1 Receptor 1 (2017) J. Virol. 91(21).
-
Hancock M. and R.L. Skalsky. Roles of non-coding RNAs in herpesvirus infection. (2017) Current Topics in Micro. and Immun., Springer Series. Ed: Ralph Tripp, Mark Tompkins
-
Skalsky R.L. Analysis of viral and cellular microRNAs in EBV-infected cells. (2017) Book Chapter: Epstein-Barr Virus, Springer Series Methods in Molecular Biology. Editors: Janos Minarovits and Hans Helmut Niller. Series Editor: John Walker
-
Skalsky R.L., Barr S.A., JefferyA.J., Blair T., Estep R., and S.W. Wong. Japanese Macaque Rhadinovirus Encodes a ViralmicroRNA Mimic of the miR-17 Family. (2016) J.Virol. pii: JVI.01123-16. *correspondingauthor
-
Skalsky R.L. and B.R. Cullen. EBV non-coding RNAs (2015) Book Chapter:Epstein Barr Virus: One Herpesvirus –Many Diseases, Springer Current Topics inMicrobiology and Immunology. Editor: Christian Munz
-
KangD., Skalsky R.L., and B.R. Cullen. EBV BART MicroRNAs Target MultiplePro-apoptotic Cellular Genes to Promote Epithelial Cell Survival. (2015) PLoS Pathogens. 11(6): e1004979.
-
Gregorovic G., Boulden E.A., Bosshard R., Elgueta Karstegl C., Skalsky R., Cullen B.R., Gujer C., Rämer P., Münz C., and Farrell P.J. Epstein-Barr Viruses(EBVs) Deficient in EBV-Encoded RNAs Have Higher Levels of Latent MembraneProtein 2 RNA Expression in Lymphoblastoid Cell Lines and Efficiently EstablishPersistent Infections in Humanized Mice (2015) J.Virol. 89(22):11711-4.
-
BogerdH.*, Skalsky R.L.*, Kennedy E.M., Furuse Y., Whisnant A.W., Flores O., Schultz K.L.W., Putnam N., Barrows N.,Sherry B., Scholle F., Garcia-Blanco M., Griffin D.E., and B.R. Cullen. Replication of many human viruses isrefractory to inhibition by endogenous cellular microRNAs (2014) J. Virol. 88(14):8065-76 *equal contributors.
-
Skalsky R.L.*, Kang D., LinnstaedtS.D., and B.R. Cullen. Evolutionary Conservation of Primate Lymphocryptovirusmicro RNA Targets (2014) J. Virol. 88(3):1617-35. *correspondingauthor
-
FloresO., Kennedy E.M., Skalsky R.L., and B.R. Cullen. Differential RISC-association of Endogenous Human microRNAs Predicts Their Inhibitory Potential (2014) Nucleic Acids Res. 42(7):4629-39
-
Skalsky R.L., Olson K.E., Blair C.D., Garcia-Blanco M.A., and B.R. Cullen. A"microRNA-like" small RNA expressed by Dengue virus? (2014) PNAS 111(23):E2359