Research
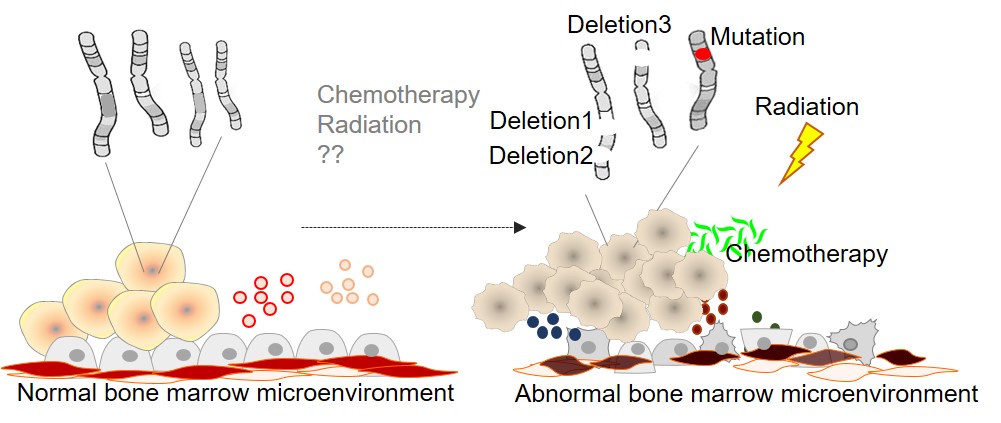
Research Focus 1: Identify and delineate the mechanisms of haploinsufficient tumor suppressor genes
Somatic genetic alterations, including mutations, translocations, and chromosome and copy number changes, are the driving force of cancer. Chromosome and copy number abnormalities account for approximately 50~60% AML, 50~80% MDS, and 25~50% MPN cases. While substantial efforts have been made in recent years to decipher the role of recurrent genetic mutations and chromosome translocations in leukemia, the pathogenesis and therapeutic vulnerabilities of chromosome and copy number aberrations remain largely uncharacterized due to the cooperation between multiple haploinsufficient genes within the deleted segments, the technical challenges of modeling these events in vitro and targeting loss of function aberrations. To address this shortcoming, our lab has recently developed and optimized a workflow that couples CRISPR/Cas screening with a sensitive cellular transformation assay to identify loss-of-function genes that yield growth factor independence and transformation of pre-leukemic cells. We have also combined CRISPR nickase with Homology DNA repair (HDR) to generate heterozygous deletions of specific genes or large chromosome segments to model clinically relevant copy number and chromosome deletions. Using these advanced models, we aim to identify novel tumor suppressor gene (TSGs) associated with chromosome and copy number deletions. The discovery and characterization of novel TSGs associated with chromosome loss and deletion will fundamentally advance our understanding of how dose perturbations of TSGs predispose to leukemia development. Ultimately, I aim to leverage these mechanistic insights to identify therapies to target the underlying dysregulated pathways of transformative cytogenetic events and ultimately improve patient outcomes.
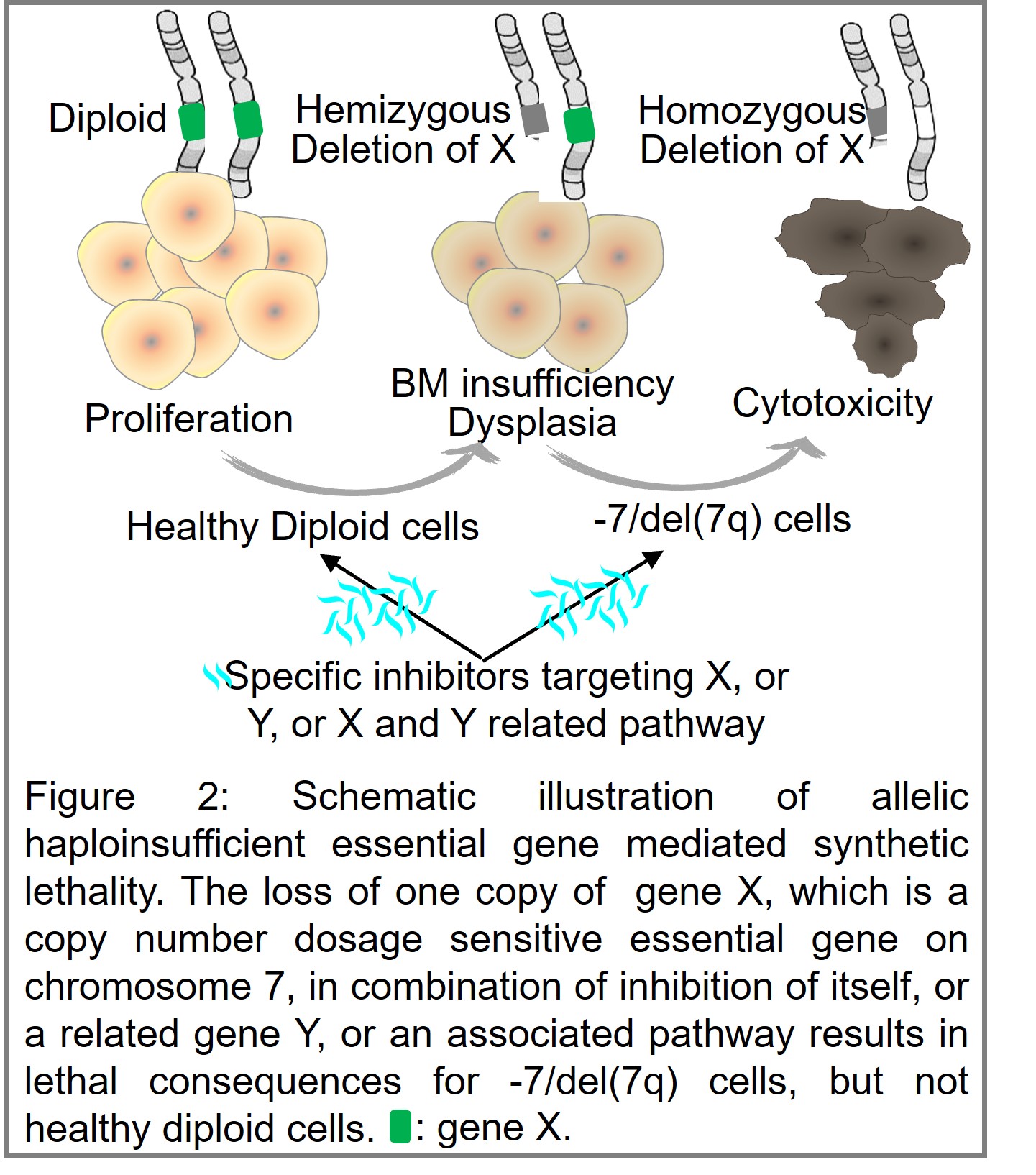
Research Focus 2: Target haploinsufficient essential genes to treat myeloid malignancies
With newly approved drugs and drug combinations adding to clinical practice, the therapeutic landscape for leukemia is changing. Despite these advances, challenges remain. Given the aggressive clinical course of leukemia, identification of biomarkers to stratify patients onto the most effective therapies and developing agents with enhanced efficacy on leukemia cells and reduced toxicity for normal tissues is an urgent need. The cellular requirement for a normal dosage of essential genes creates an opportunity to target vulnerabilities due to a reduction in the level of the protein product of a haploinsufficient gene, termed allelic haploinsufficient gene or Copy-number alterations Yielding Cancer Liabilities Owing to Partial losS (CYCLOPS) gene. Such CYCLOPS genes represent ideal biomarkers for patient stratification and synthetically lethal targets. We hypothesize there are CYCLOPS genes in the common chromosome and copy number deletion regions that could be therapeutically targeted. We will curate a list of CYCLOPS genes by analyzing gene expression, copy number, and gene dependency data from leukemia cell lines and validate clinically relevant CYCLOPS genes using shRNA and CRISPR/cas9 HDR method for future drug development.
References
Am. J. Clin Pathol 2011
Hidaka et al., Best Pract. Res. Clin. Haematol 2010
Ebert BL, Cancer Cell 2014
Ebert et al., Eur.J. Haematol 2009
Schneider et al., JCO 2008,Leukemia 2009
Sun et al., NEJM 2016
Tyner J, Nature 2008
Krönke J, Nature 2015
Papaemmanuil E, Nature 2018
Virappane P, Seifert et al.