Martin Lab

Ian Martin
Ph.D., Virginia Commonwealth University, 2008
Postdoctoral Fellow, Johns Hopkins University, 2009–2015
Faculty profile
martiia@ohsu.edu
503-494-9140
Ian received his B.S. degree from King's College, London and Ph.D. from Virginia Commonwealth University where he studied the impact of oxidative stress in determining life span and age-related functional declines in Drosophila. In 2009, he joined Ted and Valina Dawson's laboratory at Johns Hopkins University where he worked on molecular pathways underlying Parkinson's disease development linked to mutations in LRRK2 (leucine-rich repeat kinase 2). His work led to the discovery that mutant LRRK2 causes neurodegeneration through elevated bulk protein synthesis which is mediated by increased phosphorylation of the LRRK2 substrate ribosomal protein s15. In September 2015, Ian joined the Jungers Center as Associate Professor of Neurology with membership in the Parkinson Center of Oregon. Further Martin Lab information
Research
Role of aging in neurodegeneration
The biggest risk factor for developing Parkinson’s disease (PD) is age, suggesting that age-related changes in the brain predispose to loss of dopamine (DA) neuron health and viability. Cell stressors such as oxidative stress and metabolic dysfunction increase with age, and DA neurons are particularly vulnerable to this stress due to their elaborate neurite branching with consequent high metabolic demands and their intrinsic reactive oxygen species production through DA metabolism. While the case for macromolecular oxidative damage as a driver of cellular aging has recently been challenged, a growing body of evidence indicates that oxidative stress-responsive signaling may promote aging, although the molecular mediators are not well understood. We are actively investigating how oxidative stress-responsive signaling mediators that are important for lifespan determination intersect with neuronal health across age and, specifically, age-related maintenance of dopamine neuron viability.
Genetic susceptibility to neurotoxin-induced neurodegeneration
Epidemiological studies link pesticide exposure to Parkinson’s disease (PD) risk, yet human studies alone have not conclusively link PD etiology to any single pesticide. Controlled animal models play a vital role in determining whether pesticides can cause PD-related neurodegeneration and can be leveraged to gain mechanistic insight into disease etiology. We are using Drosophila to identify genes underlying susceptibility to neurotoxins such as pesticides that have been linked to PD. Genes identified through this approach will then be pursued in rodent models of neurodegeneration in order to assess their potential contribution to disease development.
Mechanisms of LRRK2-mediated neurodegeneration in Parkinson’s disease
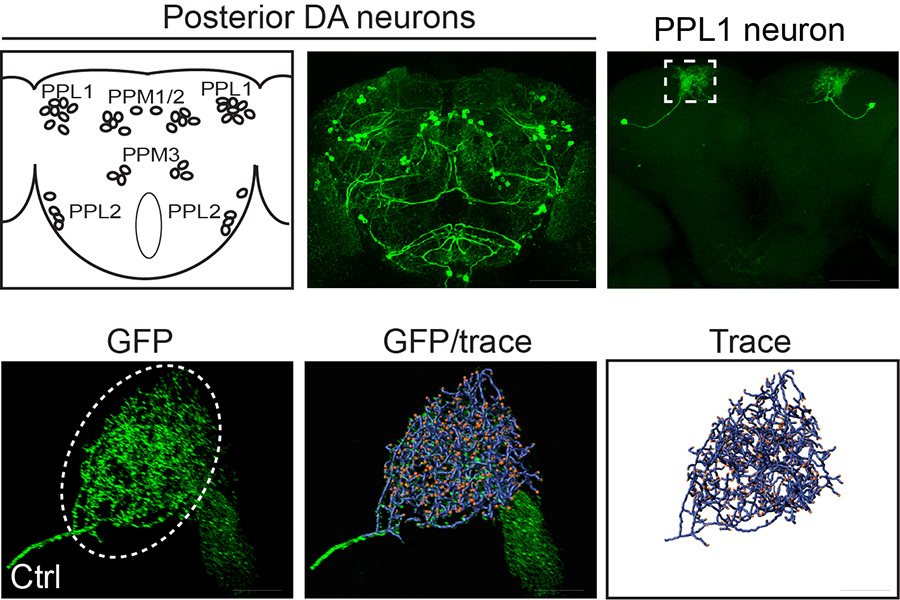
Genetic, aging, and environmental factors converge to establish a person’s lifetime risk of developing Parkinson’s disease (PD). A broad role for leucine-rich repeat kinase 2 (LRRK2) mutations in familial and idiopathic PD has emerged, elevating its status to a central therapeutic target. Ultimately, preventing LRRK2-induced neurodegeneration will require a detailed understanding of the key mechanisms driving neuronal dysfunction and death. LRRK2 has been implicated in numerous biological processes, but defining mechanisms that drive age-related neuronal death has been elusive. We performed a comprehensive screen for genetic modifiers of neurodegeneration in aged LRRK2 G2019S-expressing Drosophila, which exhibit robust age- and kinase-dependent loss of dopamine neurons (Lavoy et al., 2018). This approach identified a number of genes that regulate the morphology of neurites (axon and dendrites) which is striking considering that loss of neurite branch complexity is one of the most prevalent neuronal defects associated with pathogenic LRRK2 mutations in vitro. We are working with Drosophila and mammalian disease models to delineate the nature of these neurite defects in vivo, their underlying cause, and their relationship to dopamine neuron death.
Publications
Publications
View full list of publications on PubMed
Pallos, J., Jeng, S., McWeeney, S. and Martin, I. (2021) Dopamine Neuron-Specific LRRK2 G2019S Effects on Gene Expression Revealed by Translatome Profiling. Neurobiology of Disease, In Press.
Kim, J.K., Yin, X., Jhaldiyal, A., Kahn, M.R., Martin, I., Xie, Z., Perez-Rosello, T., Kumar, M., Abalde-Atristain, L., Xu, J., Chen, L., Eacker, S.M., Surmeier, S.M., Ingolia, N.T., Dawson, T.M. and Dawson, V.L. (2020). Defects in mRNA translation in LRRK2-mutant hiPSC-derived dopaminergic neurons leads to dysregulated calcium homeostasis. Cell Stem Cell, 27(4), 633-645.
Chittoor-Vinod, V.G., Villalobos-Cantor, S., Roshak, H., Shea, K., Abalde-Atristain, L. and Martin, I. (2020). Dietary Amino Acids Impact LRRK2-induced Neurodegeneration in Parkinson’s Disease Models. Journal of Neuroscience, 40(32), 6234-6249.
Lavoy, S., Chittoor-Vinod, V.G., Chow, C.Y. and Martin, I. (2018) Genetic Modifiers of Neurodegeneration in a Drosophila Model of Parkinson's Disease. Genetics, 209(4) 1345-1356.
Martin, I. (2017) Resveratrol for Alzheimer's disease? Science Translational Medicine 1;9(375) pii: eaam6055.
Kim, J.W., Abalde-Atristain, L., Jia, H., Martin, I., Dawson, V.L., Dawson, T.M. (2016) Protein translation in Parkinson’s disease. In: Verstreken P, eds. Parkinson’s Disease: Molecular Mechanisms Underlying Pathology. San Diego, CA: Elsevier.
Martin, I., Chittoor, V.G. (2016) Parkinson disease: Insect screens for PD therapies - keep the flies in. Nature Reviews Neurology, 12(6) 318-319.
Martin, I. (2016) Decoding Parkinson’s Disease Pathogenesis: The Role of Deregulated mRNA Translation. Journal of Parkinson’s Disease, 6(1) 17-27.
Karuppagounder, S.S., Xiong, Y., Lee, Y., Lawless, M.C., Kim, D., Nordquist, E., Martin, I., Ge, P., Brahmachari, S., Jhaldiyal, A., Kumar, M., Andrabi, S.A., Dawson, T.M., Dawson, V.L. (2016) LRRK2 G2019S transgenic mice display increased susceptibility to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-mediated neurotoxicity. Journal of Chemical Neuroanatomy, 76(Pt B) 90-97.
Martin, I., Abalde-Atristain, L., Kim, J.W., Dawson, T., Dawson, V.L. (2014) Aberrant Protein Synthesis in G2019S LRRK2 Drosophila Parkinson's Disease-Related Phenotypes. Fly, 8(3) 165-169.
Martin, I., Kim, J.W., Lee, B.D., Kang, H., Xu, J-C., Jia, H., Stankowski, J., Kim, M-S., Zhong, J., Kumar, M., Andrabi, S.A., Xiong, Y., Dickson, D.W., Wszolek, Z.K., Pandey, A., Dawson, T.M., Dawson, V.L. (2014) Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson's disease. Cell, 157(2) 472-485.
Narayanasamy, S.K., Simpson, D.C., Martin, I., Grotewiel, M., Gronert, S. (2014) Paraquat exposure and Sod2 knockdown have dissimilar impacts on the Drosophila melanogaster carbonylated protein proteome. Proteomics, 14 (21) 2566-2577.
Martin, I., Dawson, V.L., Dawson, T.M. (2011) Recent advances in the genetics of Parkinson's disease. Annual Review of Genomics and Human Genetics, vol 22 p301-325.
Martin, I., Jones, M.A., Rhodenizer, D., Alaimo, J.T., Warrick, J.M., Seroude, L. and Grotewiel, M. (2009) Sod2 knock-down in the musculature has whole organism consequences in Drosophila. Free Radical Biology and Medicine 47(6) 803-813.
Martin, I., Jones, M.A. and Grotewiel M. (2009) Manipulation of Sod1 ubiquitously, but not in nervous system or muscle, impacts age-related parameters in Drosophila. FEBS Letters 583(13) 2308-2314.
Lab members
Lab members

Judit Pallos, Ph.D.
Postdoctoral fellow
pallos@ohsu.edu
Prior to joining OHSU I studied transcriptional dysregulation in a Drosophila model of Huntington's disease at the University of California, Irvine. I enjoy working with flies because I like that we are able to use genetic and molecular biology techniques, detailed microscopical analyses, and behavioral essays all in the same model system, and get answers to our questions in a relatively short period of time. In Ian's lab I am working on two separate projects: trying to dissect the role of prospero, a transcriptional regulator, in Parkinson's disease neurodegeneration, and understanding the link between oxidative stress and aging. When not in the lab, I enjoy learning languages, playing in the kitchen, and hiking with my family.

Colin Coleman
PhD student
colemaco@ohsu.edu
Being a 3rd year Ph.D. candidate is tough, but aging is tougher. Granted, I'm only in my late 20s and therefore only experiencing it through increased heartburn and the occasional achy spine after sitting weird. However, that doesn't discount the fact that aging is the greatest environmental risk factor for Parkinson's disease, which is the main health focal point of the lab. My research interests involve finding those biological overlaps between the aging process and PD development in the hopes of discovering novel diagnostic tools or intervention strategies for the disease. Along the way, I've grown reacquainted with my old friend the mitochondria, going so far as to writing and performing an educational stand-up comedy set about them. Outside the lab, I'm a board game enthusiast, halfway-decent baker, and inconsistent cross-stitcher.

Alicia Arreola-Bustos
Undergraduate researcher
arreolab@ohsu.edu
I am currently a second year student at Portland State University pursuing a B.S. in Biochemistry with a minor in Spanish, in a pre-medical track. While in high school, I participated in several shadows at OHSU Doernbecher Children’s Hospital as well as the OHSU CHH Orthopaedic Department. I have always been drawn to medicine. I have a great appreciation for the biomedical sciences, and the application of biomedical research to patient treatment has always fascinated me. In joining the Martin Lab, I hope to learn more about the process behind research and receive experience in a lab environment. Outside of the lab, you will find me at church serving a youth group, reading, or doing some sort of project involving tons of stationery supplies.

Steffany Villalobos-Cantor
Undergraduate researcher
My interest in science and medicine has led me to pursue a degree in biology. I have previously explored this interest in diagnostic labs but it never occurred to me to delve into the world of research. I joined the Martin lab in hopes of continuing to develop lab skills and to start my career in research, and now more specifically, research of neurodegenerative diseases. My duties in the lab include maintaining Drosophila stocks, generating specific genotypes for studies that investigate the effect of diets on PD pathogenesis, and assisting with those studies. On my downtime, I enjoy running, participating in fitness classes, camping and exploring the PNW.